Reef fish timeseries
All analyses were performed in R 4.1.367. We assembled timeseries of reef fish abundance from two globally distributed databases, Reef Life Survey (RLS, https://reeflifesurvey.com/) and Reef Check (RC, https://www.reefcheck.org/), published datasets68,69 and scientific monitoring programs (Supplementary Table 7). All data consisted of quantitative surveys of reef fish abundances obtained by a combination of marine scientists and trained recreational SCUBA divers, using standardized visual methods. Methodological details, data curation and diver training are provided in refs. 70,71 for RLS and ref. 72 for RC. All surveys were conducted along transects, with the exception of data from ref. 69, which were obtained from 15-m diameter cylindrical plots. Data were aggregated by site and year by summing the abundance of individual fish species across replicate transects (and cylindrical plots), when present. We retained sites with at least five years of observation, which is appropriate for the analysis of stability and population trends in a wide range of taxa28,50,73. The final dataset consisted of 71,269 timeseries of population abundances from 2269 reef fish species sampled in 357 MPA and 747 open area sites across 50 Marine Ecoregions. Timeseries ranged from 5 to 28 years between 1992 and 2021 (February).
We identified well-enforced MPAs using the criteria set by the International Union for Conservation of Nature (IUCN)74, as areas classified either as “No-Take All” or falling in the I-III categories of protection. Expert opinions from data providers and information from published studies were used to determine the level of enforcement when IUCN categories were not applicable or the “No-Take” status was not reported. For example, Medes Islands in the Mediterranean have IUCN category V and has no reported “No-Take” status, but it is typically considered a well-enforced MPA75. Similarly, sites included in the first zoning plan of the Great Barrier Reef Park (GBRP), which was established in 1981, have neither “No-Take” status reported nor IUCN category applicable. Co-author ME distinguished between MPAs and open areas in the dataset provided for both the first and the second zoning plan, which was established in 2004. Expert opinion matched IUCN criteria for the second zoning plan, as MPA sites correspond to IUCN category II and “No-Take-All” status.
Environmental data
We considered two environmental variables as putative drivers of stability and asynchrony: marine heatwaves as an indicator of thermal stress and remoteness (the travel time to large cities) as a proxy measure of direct human pressure. Marine heatwaves were identified from daily Sea-Surface-Temperatures (SST) using the National Oceanic and Atmospheric Administration (NOAA) daily optimum interpolation gridded dataset V2.1 in the period 1 January 1982 to 31 December 202076. The dataset is a blend of observations from satellites, ships, and buoys and includes bias adjustment of satellite and ship observations to compensate for platform differences and sensor biases. Remotely sensed SSTs were obtained through the Advanced Very-High-Resolution Radiometer and interpolated daily onto a 0.25° x 0.25° spatial grid globally. Data were downloaded in January 2022 from https://www.ncei.noaa.gov/data/sea-surface-temperature-optimum-interpolation/v2.1/access/avhrr/.
A marine heatwave can be defined as an anomalously warm water event with daily SSTs exceeding the seasonally varying 90th percentile (climatological threshold) for at least 5 consecutive days36,37. The climatology was derived from the 30-yr period 1982 to 2011. We used this period as our baseline to identify marine heatwaves to comply with the recommendation of using at least 30 years for deriving a climatology, while limiting the number of instances in which the climatology extended beyond the year in which a marine heatwave was identified36. This occurred at 40 of the 1104 sites used in the alpha stability analysis and involved less than 2% of the 46,976 marine heatwave events identified in the study. Removing these sites from the analysis did not change the results (Supplementary Figure 15). The climatological mean and threshold were computed for each calendar day within a 11-day window centered on the focal calendar day across all years within the climatological period. The mean and threshold were further smoothed by applying a 31-day moving average. Two events with a break of less than 3 days were considered the same marine heatwave. All marine heatwave events were computed relative to the threshold – i.e. as the difference between the observed SST and the threshold SST. Characteristic measures of marine heatwaves, including mean, maximum, and cumulative intensity, were obtained by pairing events with fish timeseries at the site level. That is, marine heatwaves characteristics were aggregated over the same years in which fishes were sampled. We used mean marine heatwave intensity as the primary metric to quantify marine heatwaves, but we also performed a sensitivity analysis based on cumulative intensity (Supplementary Fig. 4), whereas maximum marine heatwave intensity was used to define thermal thresholds at individual sites. Marine heatwaves were identified and analyzed with the R package heatwaveR77, using SST timeseries with less than 10% of missing data.
We quantified the “remoteness” of each site as the travel time (in hours) to the closest major city (>100,000 residents), using the procedure first developed for terrestrial environments by Weiss et al.78 and adapted to marine localities by Strona et al.41. Briefly, travel time was computed from a global friction surface map (at the resolution of 1 km2) indicating the average speed at which humans can travel through each pixel using the fastest possible aquatic and terrestrial means (thus excluding aerial transportation) and then applying an algorithm to identify the least-cost path (i.e. the shortest travel time) from each site to the closest major city41.
Functional richness
Prior to analysis, species names were matched with the World Register of Marine Species79 and the FishBase80 database for validation, accessed through the R packages worrms81 and rfishbase82, respectively. We compiled six traits for each of the 2,269 fish species in the dataset, representing body size, trophic position, gregariousness, water position, intrinsic vulnerability to extinction and thermal affinity. These traits covered attributes determining species life history, trophic ecology, habitat preferences, behavior and species temperature distribution71,83. Gregariousness and water position were ordered variables, the first coded as solitary, pairing or schooling categories and the second coded as benthic (sedentary), demersal (swimming near the bottom), pelagic-reef (swimming away from the bottom within a reef) and pelagic (swimming away from the bottom among reefs) categories. The other traits were continuous variables: body size, reflecting the theoretical maximum size attainable by a species based on its growth curve; trophic position, describing the position of each species in the food web; intrinsic vulnerability, a synthetic index of the likelihood of a species to go extinct in response to fishing. Finally, thermal affinity, quantified through the Species Temperature Index (STI), measured the upper realized thermal niche of each species. This analysis required matching spatial information of species occurrences with long-term SST means. We obtained species occurrences from the Ocean Biodiversity Information System (OBIS: https://obis.org/) using the R package robis84 and SST long-term means for each occurrence location from the Bio-ORACLE v2.0 database85. To remove possible outliers, we first pruned the occurrence data by excluding extreme SST – i.e. values below the fifth and above the 95th percentiles of the temperature distribution occupied by each species. We then calculated the upper realized STI as the 95th percentile of the pruned temperature distribution of each species. All other traits were obtained from FishBase80. Continuous traits were averaged at the genus or family level for the fishes that could not be resolved at the species level (8% of the taxa); for ordinal traits, we first determined the most frequent attribute across all members within a genus or family and then converted this trait into the corresponding ordinal score.
Several functional diversity measures can be computed from a species by trait matrix33. We used the function alpha.fd.multidim in R package mFD86. The key step of the analysis is the construction of a multidimensional trait space, which is usually done through a Principal Coordinate Analysis (PCoA) applied to a Gower similarity matrix of the original species by trait matrix. Gower similarity can handle categorical, ordinal and continuous traits with missing data simultaneously, which is a desirable property for fish traits, which typically include variables of different nature, as in our analysis87. PCoA axes define a reduced multidimensional trait space within which several indices of functional community structure can be obtained at the site scale. We used the first three PCoAs in our analysis, which explained 40% of the variance on average, as a compromise between quality of trait space representation and computational speed. We focused primarily on functional richness, the proportion of the multidimensional trait space filled by all species in a site, as this measure was independent of other functional indices (correlation coefficients and 95% confidence intervals – functional richness vs. functional diversity: 0.066, 0.007–0.12; functional richness vs. functional evenness: 0.018, −0.04-0.08; functional richness vs. functional dispersion: 0.09, 0.03–0.15), which were significantly correlated (correlation coefficients and 95% confidence intervals – functional diversity vs. functional evenness: 0.15, 0.09–0.21; functional diversity vs. functional dispersion: 0.54, 0.50–0.58; functional evenness vs. functional dispersion: 0.31, 0.26–0.37). Furthermore, these alternative indices performed less well than functional richness in SEMs (see Methods: Sensitivity analyses and null models).
Stability and asynchrony
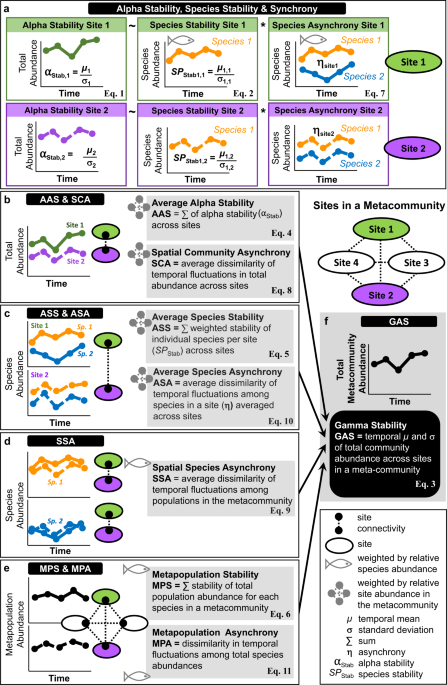
We computed six measures of stability: alpha and species stability at the site scale and gamma stability (GAS), average alpha stability (AAS), average species stability (ASS) and metapopulation stability (MPS) at the metacommunity scale5,27,32,50,88 (within ecoregions).
Alpha stability at the site scale was simply the inverse of the coefficient of variation of total fish abundance at a site:
$${\alpha }_{{Stab},i}=\frac{{\mu }_{i}}{{\sigma }_{i}}$$
(1)
where \({\mu }_{i}\) and \({\sigma }_{i}\) are the temporal mean and standard deviation of total fish abundance at site i, respectively.
Species stability at the site scale was the mean stability among species weighted by relative species abundance:
$${{SP}}_{{Stab},i}={\left(\mathop{\sum}\limits_{j(i)}\frac{{\mu }_{j(i)}}{{\mu }_{i}}\frac{{\sigma }_{j(i)}}{{\mu }_{j(i)}}\right)}^{-1}$$
(2)
where \({\mu }_{j(i)}\) and \({\sigma }_{j(i)}\) are the temporal mean and standard deviation of abundance of species j at site i, respectively.
Gamma stability was obtained as:
$${GAS}=\frac{{\mu }_{M}}{{\sigma }_{M}}$$
(3)
where \({\mu }_{M}\) and \({\sigma }_{M}\) are the temporal mean and standard deviation of total fish abundance in metacommunity M.
Average alpha stability in a metacommunity was calculated as the sum of the stability values of individual sites, weighted by relative site abundance in the metacommunity:
$${AAS}={\left(\mathop{\sum}\limits_{i}\frac{{\mu }_{i}}{{\mu }_{M}}\frac{{\sigma }_{i}}{{\mu }_{i}}\right)}^{-1}$$
(4)
Average species stability was obtained by summing the weighted stability of individual species in a site (from Eq. (2)) over sites and weighting by site relative abundance in the metacommunity:
$${ASS}={\left(\mathop{\sum}\limits_{i}\frac{{\mu }_{i}}{{\mu }_{M}}{{SP}}_{{Stab},i}\right)}^{-1}$$
(5)
where \({\mu }_{j(i)}\) and \({\sigma }_{j(i)}\) are the temporal mean and standard deviation of abundance of species j at site i.
Finally, metapopulation stability was computed as the sum of the stability of total population abundance for each species in the metacommunity, weighted by relative species abundance:
$${MPS}={\left(\mathop{\sum}\limits_{j}\frac{{\mu }_{j}}{{\mu }_{M}}\frac{{\sigma }_{j}}{{\mu }_{j}}\right)}^{-1}$$
(6)
where \({\mu }_{j}\) and \({\sigma }_{j}\) are the temporal mean and standard deviation of total abundance of species j in the metacommunity.
We computed five measures of asynchrony: species asynchrony at the site scale and spatial community asynchrony (SCA), spatial species asynchrony (SSA), average species asynchrony (ASA) and metapopulation asynchrony (MPAS) at the metacommunity scale. We first quantified synchrony using both Gross and Loreau and de Mazancourt (LdM) measures27,46,50,89 and then converted these measures into asynchrony by changing sign (Gross) or by subtracting synchrony from unity (LdM). Gross et al.46 quantified the average synchrony among species in a community as the mean correlation coefficient between the temporal abundance of each species vs. the temporal vector of the total abundance of all the other species. The index varies between −1 and 1, reflecting maximum synchrony and asynchrony, respectively, after changing sign. We used the modified version of Gross index that weights correlation coefficients by relative species abundance50:
$${\eta }_{i}=-\mathop{\sum}\limits_{j}\left[\frac{{\mu }_{j(i)}}{{\mu }_{i}}r\left({{{{{{\boldsymbol{A}}}}}}}_{j(i)},\mathop{\sum}\limits_{k\ne j}{{{{{{\boldsymbol{A}}}}}}}_{k(i)}\right)\right]$$
(7)
where \({\eta }_{i}\) is the weighted asynchrony index at site i and the term \(r({{{{{{\boldsymbol{A}}}}}}}_{j(i)},\mathop{\sum}\nolimits_{k\ne j}{{{{{{\boldsymbol{A}}}}}}}_{k(i)})\) indicates Pearson’s \(r\) correlation between the temporal vector of abundances of species j in site i (\({{{{{{\boldsymbol{A}}}}}}}_{j(i)}\)) and the vector originating from the sum of the abundances of all the remaining k species in the community (\({{{{{{\boldsymbol{A}}}}}}}_{k(i)}\)). Spatial community asynchrony quantified the average dissimilarity of temporal fluctuations among sites in a metacommunity, weighted by relative site abundance:
$${SCA}=-\mathop{\sum}\limits_{i}\left[\frac{{\mu }_{i}}{\mu }r({{{{{{\boldsymbol{A}}}}}}}_{i},\mathop{\sum}\limits_{m\ne i}{{{{{{\boldsymbol{A}}}}}}}_{m})\right]$$
(8)
where \({{{{{{\boldsymbol{A}}}}}}}_{i}\) is the temporal vector of total community abundance at site i and \(\mathop{\sum}\nolimits_{m\ne i}{{{{{{\boldsymbol{A}}}}}}}_{m}\) is the temporal vector originating from the sum of the abundances over the remaining m sites in the metacommunity. Following the same rationale, spatial species asynchrony quantified the average dissimilarity of temporal fluctuations among populations in the metacommunity, weighted by relative species abundance:
$${SSA}=-\mathop{\sum}\limits_{i}\mathop{\sum}\limits_{j}\left[\frac{{\mu }_{j(i)}}{{\mu }_{i}}\frac{{\mu }_{i}}{\mu }r({{{{{{\boldsymbol{A}}}}}}}_{j(i)},\mathop{\sum}\limits_{m\ne i}{{{{{{\boldsymbol{A}}}}}}}_{j(m)})\right]$$
(9)
were \({{{{{{\boldsymbol{A}}}}}}}_{j(i)}\) is the temporal vector of the abundance of species j in site i and \(\mathop{\sum}\nolimits_{m\ne i}{{{{{{\boldsymbol{A}}}}}}}_{j(m)}\) is the temporal vector of total fish abundance summed over the remaining m populations of species j in the metacommunity. Average species asynchrony quantified the average dissimilarity of temporal fluctuations among all species in a site (Eq. (7)) averaged among sites and weighted by relative site abundance in the metacommunity:
$${ASA}=-\mathop{\sum}\limits_{i}\left(\frac{{\mu }_{i}}{\mu }{\eta }_{i}\right)$$
(10)
Finally, metapopulation asynchrony quantified the dissimilarity in temporal fluctuations among total species abundances, weighted by relative species abundance in the metacommunity:
$${MPAS}=-\mathop{\sum }\limits_{j}\left[\frac{{\mu }_{j}}{\mu }r({{{{{{\boldsymbol{A}}}}}}}_{j},\mathop{\sum }\limits_{k\ne j}{{{{{{\boldsymbol{A}}}}}}}_{k})\right]$$
(11)
where \({{{{{{\boldsymbol{A}}}}}}}_{j}\) is the temporal vector of the total abundance of species j in the metacommunity and \({\sum }_{k\ne j} \, {{{{{{\boldsymbol{A}}}}}}}_{k}\) is the temporal vector of total fish abundance summed over the remaining k species in the metacommunity.
For comparative purposes we recalculated all asynchrony measures from 1- \(\varphi\), with \(\varphi\) indicating LdM synchrony:
$$\varphi=\frac{{\sigma }^{2}}{{\left(\mathop{\sum }\limits_{j}{\sigma }_{j}\right)}^{2}}$$
(12)
where \({\sigma }^{2}\) is the variance in total fish community abundance and \({\sigma }_{j}\) is the temporal standard deviation of abundance of species j. Equation (12) can be modified to quantify asynchrony at all the hierarchical levels addressed in Eqs. (7)–(11) (see also ref. 32). Weighted Gross asynchrony was computed using a custom function, whereas LdM asynchrony was computed using function synchrony in the R package codyn90.
Data analysis
Community-level analysis
We used Linear Mixed Effect Models to examine the relations between alpha stability (Eq. (1)), species stability (Eq. (2)), species asynchrony (Eq. (7), functional richness and their putative drivers (marine heatwaves and remoteness) and to fit piecewise Structural Equation Models (SEMs)49. All models included a random intercept for study ID, which coded for the different data sources (Supplementary Table 8) and accounted for possible generic differences in methodology among monitoring programs. In addition, we explicitly controlled for sampling effort by including the total area sampled at each site in each year as an offset in all models (see section below, Controlling for sampling effort, for details). All variables were standardized to z-scores (scaled and centered over the entire dataset) prior to analysis to provide a common scale for both responses and predictors; stability measures, remoteness and sampled area were log-transformed before standardization to improve normality. We first examined separate relationships for MPAs and open areas to match the models used in SEM, but also tested for interactions between predictors of stability, asynchrony species and functional richness and level of protection. The adequacy of model fits was assessed through a variety of diagnostics, based primarily on visual assessment of residuals using the R package performance91.
SEMs were generated separately for MPAs and open areas to reflect the hypothesized direct and indirect casual pathways among alpha and species stability, species asynchrony, functional richness, marine heatwave mean intensity and remoteness. We fitted individual pathways using the same model structure and variable transformations employed in mixed-effect models. Marine heatwaves and remoteness were exogenous variables in all models, whereas alpha stability was only an endogenous variable. All other predictors were both endogenous and exogenous variables. We started by fitting nearly-saturated global models where each endogenous variable included paths from all exogeneous variables in addition to the remaining endogenous variables, but avoiding reciprocal paths between the same variables. The only exception was the relationship between species stability and asynchrony, which was not considered since we had no a priori hypothesis about the direction of a causal path between these variables. Thus, functional richness was initially modeled as a function of marine heatwaves and remoteness; the models for species stability and asynchrony included functional richness and its predictors (marine heatwaves and remoteness); alpha stability was modeled as a function of all the other variables. We used Fisher’s C statistic to evaluate the adequacy of the global models to reproduce the hypothesized causal paths49. A model can be considered adequate when the C statistic is not significant (p > 0.05). The initial model for open areas was properly specified (Fisher’s C = 0.1, 2 d.f., p > 0.05), whereas the MPA model was not (Fisher’s C = 7.7, 2 d.f., p < 0.05). Removing the not significant link (p > 0.5) from remoteness to functional richness improved the MPA model making Fisher’s C not significant (C = 8.65, 4 d.f., p > 0.05). Results are shown as standardized effect sizes; direct and indirect effects (Fig. 3c, Supplementary Fig. 5) were extracted from SEMs using function semEff from the same R package92. Confidence intervals for standardized effects sizes were derived by nonparametric bootstrap of the fitted modes using function bootEff in package semEff.
We modeled thermal sensitivity trends using Generalized Additive Mixed Models (GAMMs) to account for the non-linear relationships between MWHs and the abundance of the four fish trophic categories. GAMMs included a tensor smooth term of marine heatwaves in interaction with MPA and open area conditions and a random smooth term for study ID. The main effect of MPA vs. open areas was evaluated in the linear part of the model. Assumptions were assessed visually by evaluating the distribution of model residuals, plots of residuals vs. fitted values and the linear predictor and plots of deviance residuals vs. theoretical quantiles. GAMMs were fitted using function gam in R package mgcv93.
Controlling for sampling effort
Our analysis required controlling for sampling effort. Although sampling methods of reef fish abundance were consistent within individual survey programs, the total area sampled varied among sites due to differences in the number of replicates and, for different programs, in the size of individual transects94. One way to account for sampling effort when investigating population trends is to divide abundance (counts) by sampled area and analyze the resulting density estimates. Unfortunately, this was not a viable approach for our analysis of stability and asynchrony because sampled area was a constant at any given site and dividing fish abundances (or any other variable) by a constant results in exactly the same values of stability and asynchrony as those obtained analyzing the original data. That the coefficient of variation – from which our measures of stability are derived – does not change when the input data are multiplied by a constant, is a well-known property of this statistic95. The same applies to measures of asynchrony since dividing timeseries of fish abundances by a constant leaves the relative differences among timeseries unchanged.
An alternative way to control for sampling effort is to include an offset in the model96. An offset is a fixed quantity associated with each observation that is used to scale the response variable, such that its influence is accounted for in the model. The offset is added to the linear predictor with a fixed coefficient of 1 (i.e. no regression coefficient is estimated for an offset) and the scaling is simply achieved by subtracting the offset from the response variable. When both the response and the offset variables are log-transformed, the scaled response variable becomes a log-response ratio (since the difference between two log-transformed quantities is equivalent to the logarithm of their ratio), which is the typical use of an offset in Poisson or binomial regression to model rates or proportions. Nevertheless, offsets can be included in other types of regression models and they are commonly employed in studies that combine data from multiple programs with varying levels of sampling effort, such as in bird surveys96,97.
Scaling the response variable by the offset requires that both variables are on the same scale. This was achieved by standardizing (i.e. scaling and centering) the response and the predictor variables, including the offset (sampled area), to z-scores. Thus, our community-level analysis shows fitted trends for scaled variables obtained as the difference between each response variable and the offset, after standardization.
Indeed, an offset may not be needed in linear models, where one could simply work with log-response ratios98. We show this equivalence in Supplementary Fig. 5, where the whole analysis is repeated by dividing each response variable by sampling effort (both log-transformed) and removing the offset from the linear model. This analysis does not assign any fixed coefficient to sampling effort, since it is now part of the response variable. Results are very similar to those obtained with an offset (e.g., the stronger negative relation between stability and marine heatwave intensity in open areas compared to MPAs). These outcomes reassure that our analysis is robust to specific choices of data transformation and that similar results are obtained whether scaled response variables are expressed as log-response ratios or as differences between standardized variables through the offset. We opted to present results based on the offset in the main text, since this improved data visualization compared to log-response ratios (compare Fig. 2b–k and Supplementary Fig. 1 with Supplementary Fig. 5).
Metacommunity networks and connectivity
We derived minimum spanning tree graphs (networks) from geographic (least-coast path distance by the sea) and biological (using Jaccard dissimilarity) distances for each metacommunity. A minimum spanning tree includes the minimum number of shortest distances to maintain all sites (nodes) connected without closed paths among nodes54. We then computed degree and closeness centrality to characterize the topology of each metacommunity network and to investigate the relationships between geographic and biological connectivity. Specifically, we employed least-squares linear regression to relate closeness centrality measured on a geographically-derived graph to degree centrality measured on a biologically-derived graph. We used functions graph.adjacency, mst, strength and closeness from package igraph99 to generate networks from distance matrices, derive minimum spanning trees and to calculate degree and closeness centrality, respectively. Degree and closeness centrality were weighted by 1/distance and scaled before analysis. Jaccard dissimilarity was computed using function vegdist in package vegan100.
Metacommunity stability and asynchrony
We compared metacommunity stability and asynchrony between MPAs and open areas within ecoregions. First, we selected ecoregions that had at least two MPA and two open area sites sampled simultaneously for at least five years. This was necessary to obtain comparable stability and asynchrony measures. There were 12 ecoregions that met these criteria. Since there were many possible ways to combine sites and years, we developed an algorithm to select the combination of years that maximized the number of MPA and open area sites (an alternative algorithm that maximized the length of matching timeseries yielded too few sites in most ecoregions).
Second, we calculated a matrix of least-cost path distances by the sea (i.e. avoiding land masses) among the selected sites for each of the 12 metacommunities using function costDistance from terra package in R101. We used these distance matrices to match MPA and open area sites within the spatial scale defined by the maximum distance separating any two MPA sites within an ecoregion. For each MPA site we identified all other MPA and open area sites within the defined spatial scale and computed all metacommunity stability and asynchrony measures from these sites. This procedure was repeated for all MPAs in a metacommunity and the results were averaged. Inevitably, all MPA sites became selected at each iteration (they were all included within their maximum distance, by definition), thus, only the stability and asynchrony measures obtained from one iteration were retained for MPAs. As a sensitivity test, we repeated the analysis by matching MPA and open area sites within a spatial scale of 50-100 km, which was intermediate between the maximum distances separating MPAs in metacommunities (Supplementary Table 5), with 100 km representing a potential upper limit of direct fish dispersal40,56. Although our matching procedure used the same sites more than once, averages were independent between MPAs and open areas.
Third, we compared the stability and asynchrony measures between MPAs and open areas within each of the spatial scales defined above. To do so, we developed a simulation approach to obtain robust estimates of variances for each stability and asynchrony measure and level of protection. Although we had access to primary fish abundance data, Eqs. (2)–(6) and Eqs. (8)–(11) necessarily generated a single value for each metacommunity precluding the direct estimation of variances. We addressed this problem through a Jacknife (leave-one-out) simulation approach, which consisted in recalculating all stability and asynchrony measures for each metacommunity by excluding one species at the time. The resulting variances were used to derive the Hedge’s g effect size of the difference between MPAs and open areas for each measure, which we analyzed in a Bayesian meta-analytical framework. We used a model of the following form:
$${y}_{m}={gaussian}({\theta }_{m},\,{\sigma }_{m}^{2})$$
(13a)
$${\theta }_{m}={gaussian}(\mu,\,\tau )$$
(13b)
where \({y}_{m}\) was the estimated Hedge’s g effect size for any of the measures analyzed in metacommunity m, which was assumed to originate from a Normal distribution centered on the true effect size \({\theta }_{m}\) with variance \({\sigma }_{m}^{2}\). Metacommunity m was considered a random sample from a population of possible metacommunities, such that \({\theta }_{m}\) itself originated from a Normal distribution with true mean \(\mu\) (the true population-level effect size) and dispersion parameter \(\tau\). We used weakly informative priors for parameters (a normal priori for \(\mu\) and a Cauchy prior for \(\tau\)):
$$\Pr \left(\mu \right)=N\left(0,\,1\right)$$
(13c)
$$\Pr \left(\tau \right)={Cauchy}\left(0,\,1\right)$$
(13d)
Separate models were fitted for each metacommunity stability and asynchrony measure using function brm from the brms R package102. Models run for 4000 iterations, 1000 burn-in iterations and 4 chains; other tunable parameters in brm function were left to their default value. Model convergence was assessed through visual inspection of trace plots and ensuring that the \(\hat{{{{{{\rm{R}}}}}}}\) parameter – a key diagnostic of convergence – was equal to unity. Since gamma stability was examined within metacommunities, which generally included data from individual programs with consistent methods and sampling effort, an offset was not included in these analyses. Finally, we examined the relations between posterior distributions and three attributes of MPA networks, spatial scale, number of MPAs and number of sampled sites, through linear regression.
Sensitivity analyses and null models
We performed a series of additional tests to evaluate the sensitivity of our results to analytical detail and methodological differences among monitoring programs. Checks were particularly needed for community-level analyses, which compared data across monitoring programs. We assessed the robustness of results and conclusions from the analysis of alpha stability to specific choices of asynchrony (Gross vs. LdM) and functional measures (richness, diversity, evenness, dispersion) and to detrending of timeseries47. All functional measures were weakly associated with alpha stability and asynchrony, but functional richness had stronger path coefficients than the other functional measures in SEMs and resulted in a lower Fisher’s C score contributing to a better representation of the hypothesized casual pathways.
Monitoring programs differed in their taxonomic scope: although most of them were designed to survey all the species occurring in sampling units, some targeted a pre-determined subset of the species (e.g., RC). We performed two analyses to evaluate whether differences in taxonomic scope and other methodological details among monitoring programs affected the main results. First, we evaluated the robustness of one key result, the positive effect of MPAs on alpha and species stability, asynchrony and functional richness with intensifying marine heatwaves, by excluding study IDs with 50 species or less (Supplementary Fig. 6). Second, we used sample coverage48 to evaluate whether fish communities were adequately sampled in MPAs and open areas, regardless of differences in taxonomic scope, sampling effort and size of sampling units (transects, cylindrical plots) among monitoring programs. Sample coverage is a measure of sample completeness and gives the proportion of the total number of individuals in the community that belong to the species represented in a sample of that community. Sample coverage can be calculated by rarefying (subsampling) the community, or by extrapolating abundance or incidence data to a pre-determined value (typically, twice the total observed abundance or number of samples)48. Subtracting sampling coverage from unit gives the “coverage deficit”, the probability that a newly added individual (for abundance data) or sampling unit (for incidence data) belongs to a previously unseen species in the sample. We compared sample coverage between MPAs and open areas for different size categories of sampling units (transects or cylindrical plots) using function iNEXT in the same R package103.
Although most programs started after the enforcement of protection, some (e.g., the GBRP) embraced both before and after periods. These timeseries could include spikes of fish abundance and diversity in response to protection that may not have occurred in timeseries including only after data, with unknown consequences on estimates of stability and asynchrony. To assess this potential bias, we repeated the analysis of alpha stability by excluding data sampled before the establishment of an MPA from those timeseries that encompassed both periods. Results were qualitatively similar to those of the main analysis and are thus not reported here.
Finally, we ran null models to assess whether fish species fluctuated more or less asynchronously than expected by chance in metacommunities. Null models consisted of 999 iterations of the cyclic shift algorithm, a common method to preserve temporal autocorrelation in simulated timeseries. We applied the cyclic shift algorithm independently to each individual species at a site. Observed timeseries were considered one realization of the null model and were combined with those originated by the cyclic shift algorithm, using the function cyclic_shift in R package codyn90.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.